The primary objective of research in our lab is to dissect the fundamental molecular mechanisms governing the role of protein post-translational modifications in epigenetic and chromatin regulation and signaling pathways required to respond to environmental cues. We are currently interested in methylation of histones and other proteins, a modification system that has been well-established to regulate chromatin functions and diverse signaling pathways. Aberrant regulation of lysine methylation disrupts chromatin homeostasis and has been implicated in diverse human pathologies, particularly cancer, neurodevelopmental disorders, and neurodegenerative diseases.
Our research uses the model system budding yeast and mammalian cell lines to address fundamental biological questions and uncover molecular pathways that are disrupted in disease. Three ongoing areas of research in the lab are described below:
Calibrating gene expression programs during stress through chromatin regulation

Chromatin-modifying and remodeling factors are key drivers of genome reprogramming in response to fluctuating environmental signals and during stress. The misregulation of chromatin factors is a major mechanism underlying diseases such as cancer and neurological disorders, aberrant development, and aging. However, in many cases, the mechanisms directing chromatin regulation in response to stress to precisely calibrate gene expression are not understood. A major focus of research in our lab is to dissect the biological function of a group of noncanonical SET domain proteins that regulate gene expression and chromatin function and are clearly implicated in multiple cancer types and neurodevelopmental disorders. To this end, we have been investigating the molecular functions of the budding yeast protein Set4 and have demonstrated that it plays important roles in gene regulation in response to oxidative stress and hypoxia. We are currently expanding these studies to better define the functions for related proteins in yeast and mammalian cells, particularly in response to stress, and to understand their roles in disease. Some of our key publications on this topic include Tran et al., J Biol Chem, 2018 and Jethmalani et al., Life Sci Alliance, 2021. Our reviews highlighting the biological importance and molecular mechanisms of these proteins include Tran and Green, Curr Genet, 2019 and Sun et al., J Mol Biol, 2024.
Defining SMYD protein activity and functions in chromatin and signaling
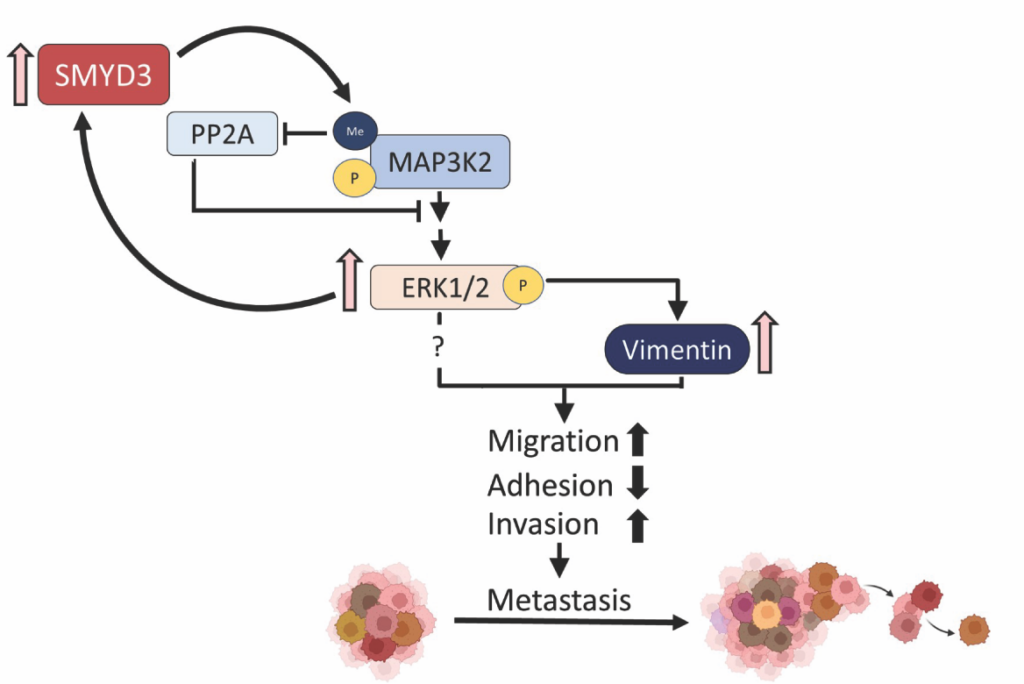
SMYD enzymes, a distinct subfamily of SET domain-containing lysine methyltransferases, are well-conserved and present in model systems (two in budding yeast) and humans (five in humans). Mammalian SMYD enzymes have been extensively studied for their contribution to oncogenesis, and are also key regulators of skeletal and cardiac muscle development and differentiation. Altered SMYD enzyme levels have been linked to age-related pathologies including muscle atrophy and cardiac disease and they are frequently dysregulated in cancer. Biochemically, SMYD enzymes appear to primarily target non-histone proteins for methylation though may also have some role in regulating chromatin via methylation. However, our understanding of their biochemical activities and functions is still relatively limited. We have worked to characterize the budding yeast SMYD enzymes, with a focus on yeast Set5 in published work. In human cells, we have also characterized the molecular role for SMYD3 in prostate cancer metastasis by linking its function to MAP3K2/MEK/ERK signaling. These studies are currently being expanded to better understand some of the fundamental functions of SMYD enzymes by further defining their roles in yeast and further our molecular investigation of SMYD enzyme function in prostate cancer and other solid tumors. Some of our key publications on yeast Set5 include Green et al., Nat Struct Mol Biol, 2012 and Jaiswal et al., Mol Cell Biol, 2020. Key publications on human SMYD3 include Ikram et al., Sci Adv, 2024. We have also reviewed the topic of protein methylation in Jethmalani et al, Curr Prot Pept Sci, 2020.
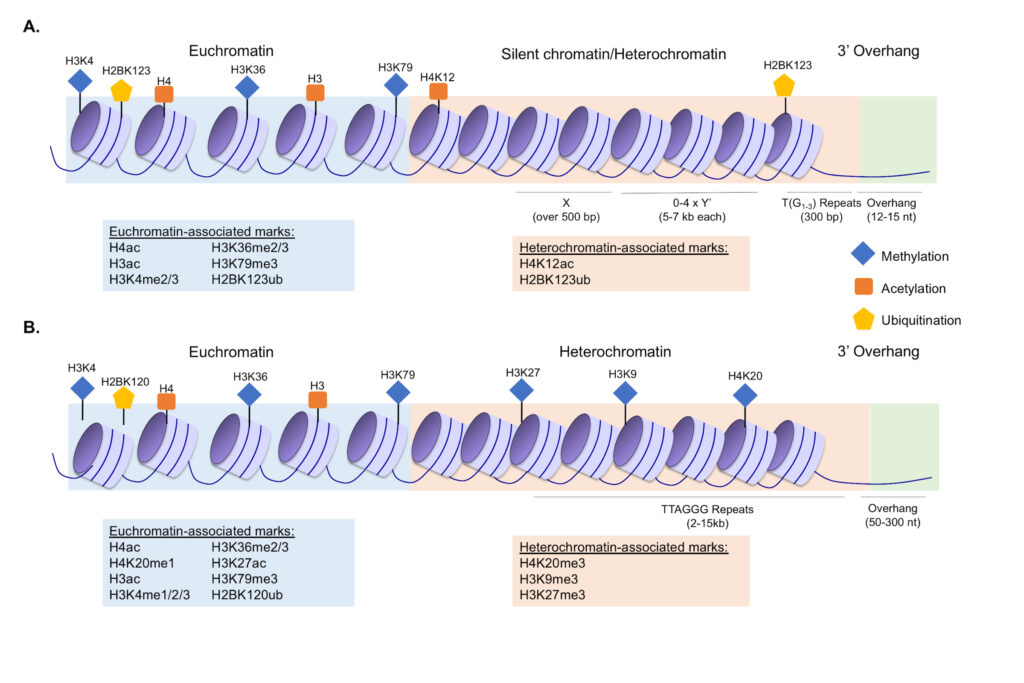
Understanding the role for histone methylation in telomere maintenance
The highly-conserved H3K4 methyltransferase Set1 is linked to both active transcription and gene repression, however there are many unanswered questions regarding how its diverse functions regulate the genome. The loss of Set1 has long-been linked to defects in subtelomeric chromatin, however the specific role for Set1 in this process is still unclear. We investigated numerous proposed hypotheses regarding the role for Set1 and H3K4 methylation in telomere regulation, which led us to discover that multiple telomere maintenance factors rely on Set1 to properly calibrate their abundance in the cell through transcriptional, post-transcriptional, and post-translational mechanisms. This work has shed light on mechanisms of telomere regulation in yeast and new functions for Set1 and H3 K4 methylation. Furthermore, despite extensive links between human orthologs of Set1 and pathologies of telomere misregulation including cancer and aging-associated disorders, there has been very little molecular investigation of the role for Set1 and H3K4 methylation in telomere maintenance in humans. Some of our key publications on this topic include Jezek et al., Epigenetics, 2017 and Jezek et al., Mol Biol Cell, 2023. We have also reviewed the general role for histone modifications in telomere maintenance in Jezek and Green, Cells, 2019.